ProteoWizard is a set of open-source tools and libraries for proteomics researchers and includes file conversion tools that allow vendor-specific file formats to be converted to standard formats (mgf, mzML, mzXML and mzData). Learn how to download and install ProteoWizard here: http://proteowizard.sourceforge.net/projects.html
Convert TripleTOF system *.wiff files to mzML
Recommended settings for converting TripleTOF system files to mzML: Ensure that the “Peak Picking” filter is the very first filter in the list. Otherwise, it will cause the ProteoWizard software to ignore the vendor peak picking and it might get the wrong mass for most peaks.
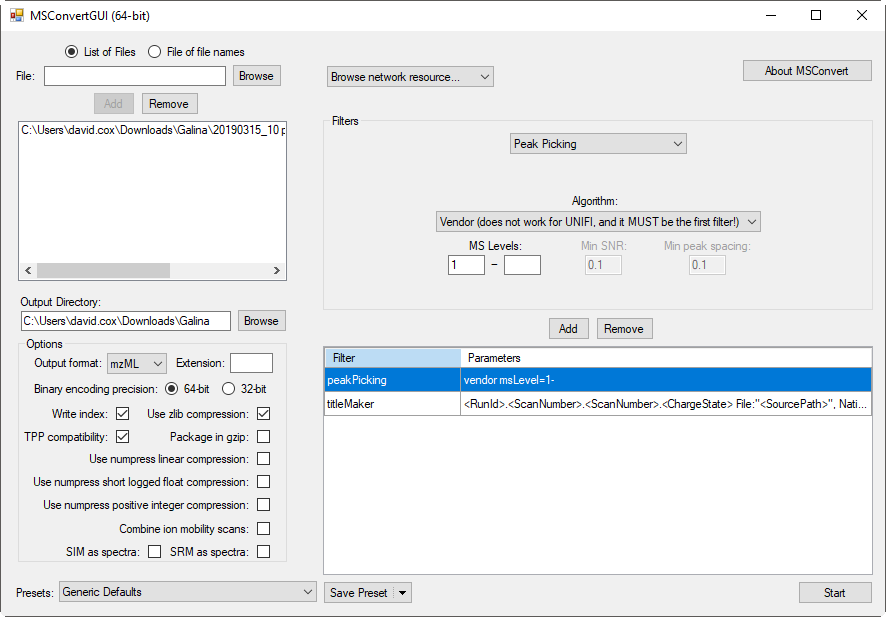
Note that Proteowizard now writes out the values by peak area not peak height. Also they have recently adding a scaling factor of 100 to the peak area values as some 3rd party software cannot handle values below 1 for peak area. So please make sure you are using the latest ProteoWizard version.
Converting Thermo Q Exactive files for use in ProteinPilot software
ProteinPilot software can process MS data from any vendor as long as the files are converted into mgf format. ProteoWizard file conversion tools enable vendor-specific file formats to be converted to the standard mgf format.
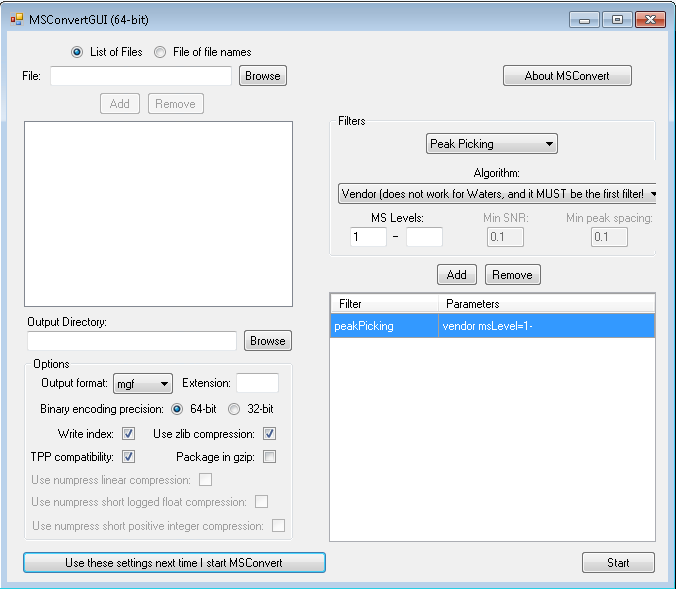
Below are some good starting settings to convert Q Exactive files to mgf using ProteoWizard, for searching with ProteinPilot software:



 Contact Support
Contact Support
0 Comments