Science and Technology Award – HUPO 2017
 We are pleased to congratulate its research scientists Stephen Tate and Ron Bonner (retired) for being awarded this year’s Science and Technology award at HUPO 2017 in Dublin Ireland. The Science and Technology Award at HUPO recognizes an individual or team who were key in the commercialization of a technology, product, or procedure that advances proteomics research.
We are pleased to congratulate its research scientists Stephen Tate and Ron Bonner (retired) for being awarded this year’s Science and Technology award at HUPO 2017 in Dublin Ireland. The Science and Technology Award at HUPO recognizes an individual or team who were key in the commercialization of a technology, product, or procedure that advances proteomics research.
I sat down with Steve and Christie recently to chat about the HUPO award and the evolution of SWATH® Acquisition, from its early days as a technology innovation to today’s broad adoption.
Tom: Can you talk a little bit about how the concept of SWATH Acquisition came about?
Steve: Back in 2008-2009, the main quantitative techniques used for proteomics research were either label free, label based or spectral counting using Data Dependent Acquisition (DDA) or Multiple Reaction Monitoring (MRM). MRM quantitation was becoming extremely popular at this time as it overcame some of the issues of data completeness, long run times and inconsistent quantitation which plagued the DDA methods. The MRM based techniques provide high quality quant and good throughput but were limited in the number of proteins that could be monitored and required in-depth method development time. Although the benefits outweighed the development time, the fact was that the desire to perform systems biology level analysis by Mass Spectrometry on a scale similar to that which was being undertaken by genomics methods required a different technique.
So this got us thinking about how we could have higher quality quantitation, equivalent to that of MRM but with significantly less assay development, a sort of a “Lazy person’s MRM quantitation.” At the same time, we were developing the next generation QTOF – the TripleTOF® platform, which had significant improvements in sensitivity and speed, allowing us to try novel methods for analysis. These methods led us to the SWATH technique.
The first was MS/MSALL, where we stepped a 1Da quad isolation window across the mass range collecting full scan MS/MS data. Although this was not new and had been around on the QSTAR® system, the technical innovations in the TripleTOF platform resulted in this being a useful scan that has significantly utilized in untargeted lipidomics.
The second was method was a derivative of the MRM workflow, where the instrument selects the mass of individual peptides and the full scan MS/MS data acquired from the TOF platform provided a rich source of data. The MRMHR workflow as it became known provides a method for targeting a significant number of peptides with the specificity of MRM and sensitivity of a triple quadrupole MS system. It also allowed a large number of peptides to be monitored, but still, there was a desire to increase this number further.
In late 2009 Ruedi Aebersold visited SCIEX in Toronto, where we talked to him about the upcoming TripleTOF system. I remember the meeting vividly as Ruedi had literally just landed prior to heading to the office and we presented all of the novel technology in the platform and a lot of the collected data. We also included a few slides on technology changes which were not yet utilized in the methods, one being the ability to isolate a wide range of mass with great fidelity, LC time scale MS/MSALL essentially. Ruedi was excited by this aspect and we had a long discussion on how to marry the wide band pass isolation, the peptide spectral libraries that his team had been developing for MRM assay development, as well as potentially some of the automated scoring methods. As a result of these discussions, we realized that putting the technology together with the libraries and algorithms could be a significant workflow innovation and potentially a method which would have the same characteristics of MRM but on a complete proteome scale.
Tom: What were the technical challenges in making this new protein quantitation solution a reality?
Steve: Well, there were really three main innovations that had to come together to make this a viable solution and product.
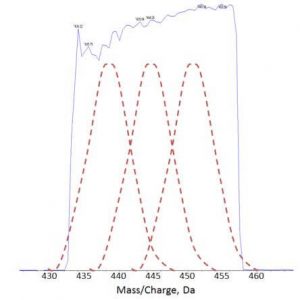 Technically we had to overcome the time taken to accumulate ions for each mass so that it could “fit” with the LC-time scale allowing quantitation. This tradeoff between accumulation time and peak width limited the MS/MSALL scan to essentially an infusion technique. In overcoming this, we first had to isolate a wider mass range in the quadrupole. However normally at this time, the isolation efficiency diminished as you moved to the edges which caused reductions in the signal and limited the quantitation ability (red dotted lines in fig). The ability to generate square quadrupole isolation windows of wider width was a result of the work of our R&D team. The redeveloping of the electronic control of quadrupoles and also developing the mathematics to allow for the control resulted in increased resolution of the isolation boundaries and squarer isolation width – both critical for the quant (blue line in fig).
Technically we had to overcome the time taken to accumulate ions for each mass so that it could “fit” with the LC-time scale allowing quantitation. This tradeoff between accumulation time and peak width limited the MS/MSALL scan to essentially an infusion technique. In overcoming this, we first had to isolate a wider mass range in the quadrupole. However normally at this time, the isolation efficiency diminished as you moved to the edges which caused reductions in the signal and limited the quantitation ability (red dotted lines in fig). The ability to generate square quadrupole isolation windows of wider width was a result of the work of our R&D team. The redeveloping of the electronic control of quadrupoles and also developing the mathematics to allow for the control resulted in increased resolution of the isolation boundaries and squarer isolation width – both critical for the quant (blue line in fig).
The advances in QTOF technology were also critical. The scan speed increases of Triple TOF technology allowed us to perform a high number of looped scans in a single experiment which when coupled with the wide band pass filtering in the Q1 quadrupole allowed a complete sweep of the mass range of interest, and in a time that allowed enough data points to be collected across an LC peak. The increase in the spectral resolution in the TripleTOF platform also allowed us to in silico extract the exact masses with high confidence in the data. Really without any of these components, it would have been impossible to perform SWATH acquisition with the degree of reproducibility that we have seen and still strive to push further.
On the software side, the challenge was how we were going to deconvolute this data to produce searchable data to push into the protein identification workflow. Well, the answer was we don’t. The workflows for the identification of peptides and proteins are well established, and our goal was for SWATH acquisition to be a quantitative workflow. The targeted extraction proved to be the key concept here to make this a reality, combining the known information present in empirically generated peptide spectral libraries, such as those generated by the Aebersold group. This complex data set provided a first glimpse of the potential power of this method for protein quantitation at large scale. Reducing this data to MRM like signals and processing the data from a quantitative standpoint using established methods allowed for and simplified processing of this highly dimensional data set. Leveraging both the full scan and MRM-like aspects of the data, along with the chromatographic information in the scoring process allowed a high confidence in the final results.
It was truly a cross-functional team that pushed this through, and a significant partnership with the Aebersold group. Put this together, and you have the first SWATH acquisition workflow, as presented by Dr. Ruedi Aebersold at HUPO 2010 in Sydney Australia!
Tom: How has SWATH Acquisition evolved since its inception?
Christie: We have been thinking a lot about this lately since we recently celebrated the 5 year anniversary of SWATH at ASMS this year (2017). When we first introduced SWATH acquisition at HUPO 2010, then made it commercially available to researchers at ASMS 2012, the acquisition strategy was using 32 Q1 isolation windows (25 Da each) to cover the peptide m/z range in an LC compatible time frame. We realized pretty quickly that our data was limited by the S/N and started investigating moving to smaller Q1 windows, and of course, the speed of the TripleTOF system let us evaluate this impact. The team explored a number of different methods for running windowed isolation including multiplexed windows, variable windows and of course the MS/MSALL 1amu windows. The introduction of the variable window SWATH technique enabled us to build methods with much smaller isolation windows in an optimized fashion according to the m/z composition of a sample, allowing us to extract significantly more data from our SWATH acquisition experiments. Most recently, we have also scaled up our SWATH experiments, moving the LC to microflow rates to improve throughput and robustness. Learn more about the evolution of SWATH Acquisition >
There has also been a lot of progress made around data processing by the SWATH acquisition user community! Algorithms enabling identification of proteins directly from SWATH acquisition data have emerged and are showing promise. Numerous other software tools have emerged enabling the processing of SWATH acquisition data for other workflows.
Tom: How has SWATH Acquisition changed proteomics?
Christie: The evolution and uptake of SWATH acquisition has been remarkable.
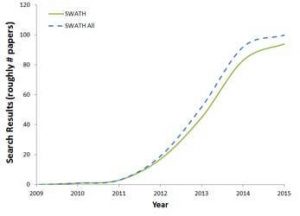 The list of applications and publications that use SWATH acquisition is extensive and continues to grow every month. A quick search with Google Scholar (figure) gives a sense of the breadth and promise of this exciting MS technique as it continues to develop and make inroads into new areas of research. We are seeing the use of SWATH acquisition in high impact papers in Science and Cell, suggesting this is now a mainstream technique.
The list of applications and publications that use SWATH acquisition is extensive and continues to grow every month. A quick search with Google Scholar (figure) gives a sense of the breadth and promise of this exciting MS technique as it continues to develop and make inroads into new areas of research. We are seeing the use of SWATH acquisition in high impact papers in Science and Cell, suggesting this is now a mainstream technique.
The transition from traditional proteomics workflows to SWATH Acquisition has transformed proteomics experiments, allowing data to be robustly captured across very large sample cohorts. This combined with other growing trends in data acquisition, such as higher-throughput microflow chromatography have unleashed a new era of more industrialized proteomics. This can be clearly seen through the opening of the large biomarker discovery centers in recent years, such as the Stoller Centre (Manchester, UK) and ProCan (Sydney, Australia). These centers have implemented microflow SWATH Acquisition to industrialize their approach to proteomics, such that they can analyze large sample sets with the goal of advancing Precision Medicine. The success of Precision Medicine depends on a repository of biological knowledge that reflects the diversity of the population. This means measuring more samples and larger cohorts. SWATH Acquisition is proving to be an excellent strategy to achieve this!
Tom: Where do you see SWATH Acquisition going in the future?
Christie: SWATH acquisition has really had a significant impact on the field of proteomics and is now set to revolutionize other fields.For example, food-testing laboratories survey products to test for key ingredients and contaminants, to ensure food quality and safety. Pesticide residues in certain foods such as baby food are required to be very low, and foods such as fruits or vegetables often exhibit a high number of active components. SWATH Acquisition allows scientists to completely survey samples for every detectable chemical contaminant present while also ensuring rapid, reliable detection of low abundance compounds.
The challenge of forensic testing involves routine screening and quantification of drugs and medications in numerous biological matrices, such as blood and urine. SWATH technology collects comprehensive MS data and MS/MS data from a single sample injection, which results in a digital archive of the forensic sample. This allows re-interrogation of the sample without physical re-analysis, removing the need for expensive sample storage and saves time. This provides high quality results that will stand up in court.
In life sciences research, the scale and depth of data that can be achieved with SWATH acquisition will be key in developing comprehensive views of biology and disease. We are already starting to see trends around generating these larger sample sets with an eye on digital biobanking. Also, the increased use of SWATH acquisition in other omics such as metabolomics will enable a true multi-omics approach, where comprehensive, quantitative “Big Data” can be acquired in areas such as systems biology and precision medicine.Learn More in SWATH Discussions >






 Contact Support
Contact Support
0 Comments