To perform automatic mass recalibration, ProteinPilot software first conducts a rapid search using mass tolerances that are wider than expected for the instrument in use. The mass tolerance used in this initial search is defined in the MSTOLERANCE VALUE and MSMSTOLERANCE VALUE settings in the parameter translation file (Figure 1). To see what these values are for your instrument, refer to your parameter translation file (located at C:\Program Files\AB SCIEX\ProteinPilot\WorkflowDirectory\ParameterTranslation.xml).
<INSTRUMENT xml:id=”INSTRUMENT:62″ name=”TripleTOF 6600″ ionization=”ESI” taglet_ef=”EF_CURVE:1″ fraglet_correction=”1″ quant_threshold_sn=”5″>
<AUTO_CALIBRATION ms=”true” msms=”true” msmstolsdratio=”2.75″ mstolsdratio=”7″ type=”dalton” squareroot=”true”/>
<MSTOLERANCE VALUE=”0.05″ TYPE=”dalton”/>
<MSMSTOLERANCE VALUE=”0.1″ TYPE=”dalton”/>
<MS_STANDARD_DEVIATION VALUE=”0.0011″ TYPE=”dalton”/>
<MSMS_STANDARD_DEVIATION VALUE=”0.01″ TYPE=”dalton”/>
<DEFAULT_CHARGE_STATE charge=”2″/>
<DEFAULT_CHARGE_STATE charge=”3″/>
<DEFAULT_CHARGE_STATE charge=”4″/>
</INSTRUMENT>Figure 1. The parameter translation file defines the mass tolerance values and various other settings for the instrument in use.
The high-confidence identifications (of non-modified tryptic peptides) from this quick search can then be used to determine the mass shift and compute a mass correction. The correction is performed in segments across the duration of the LC run, just in case there is minor drift across the run time (binned by every 4,000 MS/MS spectra). For both the MS and MS/MS data, each segment is individually recalibrated. To compute the calibration within each RT range, the m/z range is binned and the distribution of the delta between theoretical and observed m/z in each bin is evaluated for the mode location. Predefined m/z tolerances around the mode of the delta in each m/z bin are used to reject outliers from the analysis. A linear regression is done in the TOF (sqrt(m/z)) domain.
The resulting recalibrated peak list can be used for the final search along with tighter mass accuracy settings that are appropriate for a well-calibrated instrument of your instrument type (Figure 2). These tolerances are defined in the parameter translation file as MS_STANDARD_DEVIATION VALUE and MSMS_STANDARD_DEVIATION VALUE (Figure 1), and they reflect the typical mass variation expected for each instrument. The final mass errors that are reported are based on the recalibrated results.
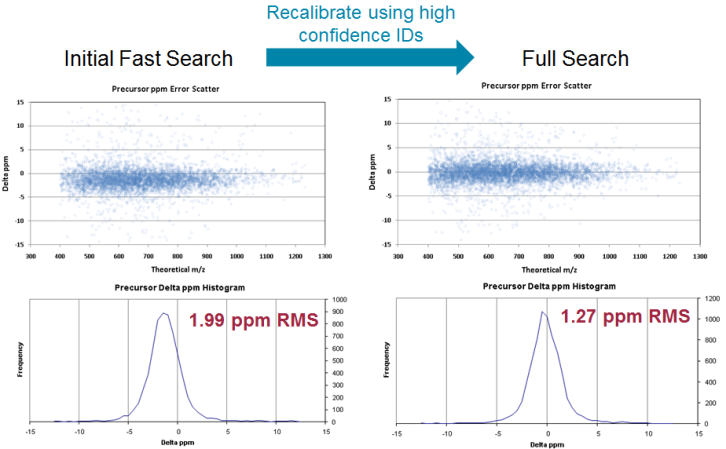
Figure 2. The peak list that results from the recalibration can be used for a full search along with tighter mass accuracy settings appropriate for a well-calibrated instrument.
Note that the calibration is performed separately for MS and MS/MS experiments.
Also note that with ProteinPilot software 5.0, you can export the recalibrated peak list as an *.mgf file and use it to compare the search results with results from other search engines.
For advanced users: Interested in investigating the impact of mass recalibration? Although we recommend always using mass recalibration, the algorithm can be turned off if you want to investigate its impact on your results. To turn off the mass recalibration algorithm:
- Close the ProteinPilot software, open the parameter translation file (Figure 1) and edit the file as described in the next step.
- Find the instrument definition for your TripleTOF system type, find the AUTO_CALIBRATION flag and change the flag from True to False to turn off recalibration.
- After you have performed your search, remember to turn calibration on again by repeating steps 1 and 2 (this time, changing the flag from False to True).



 Contact Support
Contact Support
0 Comments