RNA experiments can be created in the OneOmics suite for multi-omics analyses, enabling integration of transcriptomics data and proteomics data for biological insight. To build RNA experiments, either CloudConnect for PeakView software 2.2 or BaseSpace (Illumina) can be used to upload transcriptomics data files for use within the SCIEX Cloud Platform.
If using BaseSpace, ensure that your BaseSpace account is linked to your SCIEX Cloud account before beginning the upload by clicking the settings icon in the upper right-hand corner of the home page, and then navigating to Connect Services. To link to your account in BaseSpace, select Connect under the BaseSpace options and this will bring up the normal login page for BaseSpace. Enter your email address and password.

CloudConnect can be used to upload RNA-sequencing data files directly to Data Store within the SCIEX Cloud Platform. Simply select the files you wish to upload and specify a folder for storage. After uploading, the files will be available for further processing in the OneOmics suite in the specified folder.
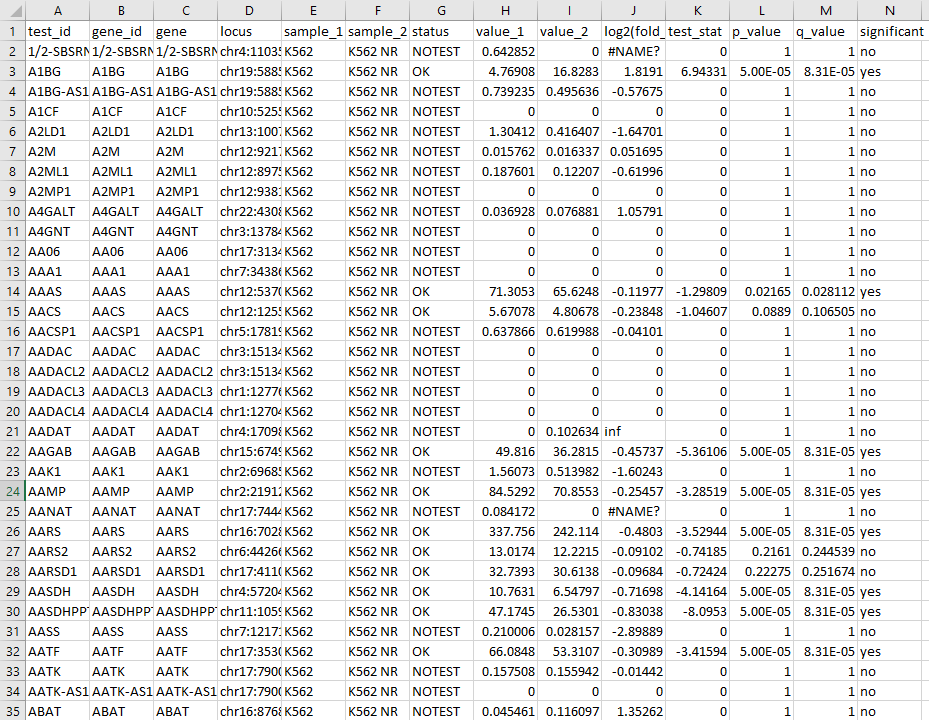
Please note that there are two file types that can be used to build RNA experiments—either gene_exp.diff files or .txt files. For text files, the RNA-Seq data in the file must have the same columns as the standard CuffDiff format, with the correct headers. All columns must contain values, but only the values in the following four columns are imported: Gene, p_value, log2(fold_change), and status. The gene names must be provided as HGNC-approved gene symbols. Additionally, only rows with the Status value ‘OK’ (test successful) are imported. An example of the correct file input structure can be found in the Data Store, in the Shared by SCIEX > Getting Started > Multi-Omics > TKI-Resistance RNA-Seq folder.
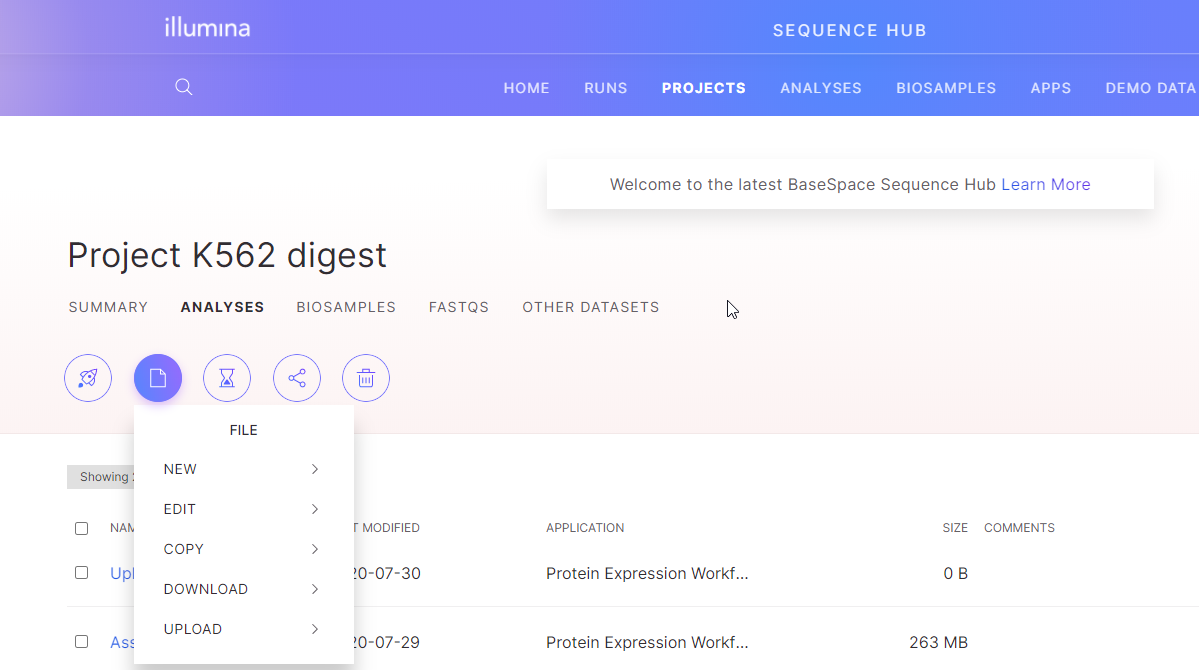
In BaseSpace, you can build projects containing your transcriptomics data files and, for RNA experiments, these can either be CuffDiff or text files. Create a new project folder in BaseSpace, and then within the project folder. You can select the paper icon to upload files to your project. Your project names in BaseSpace will be viewable in SCIEX Cloud and you can access the files within to build your experiments in Experiment Manager.
Once you have uploaded your files, navigate back to the OneOmics suite to build your RNA experiment in Experiment Manager.
In Experiment Manager, create a new RNA experiment and then navigate to either the Data Store or BaseSpace folder that contains your gene_exp diff files or text files. Select the files of interest, and then click Create to generate the experiment.
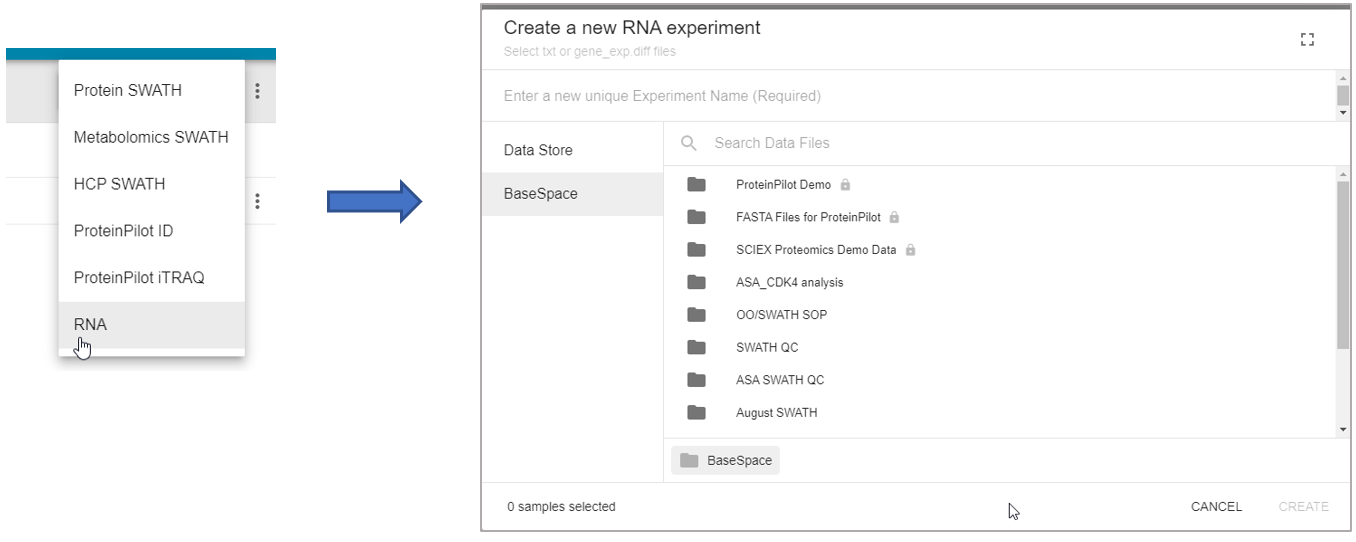
Next, you will need to categorize your gene_exp diff files or text files according to Experiment Group and Control Group. Once the metadata has been put in, a green checkmark will appear by your RNA experiment and it can be saved for further processing in the OneOmics suite. Please see the Cloud Talk for more information on building Experiments.
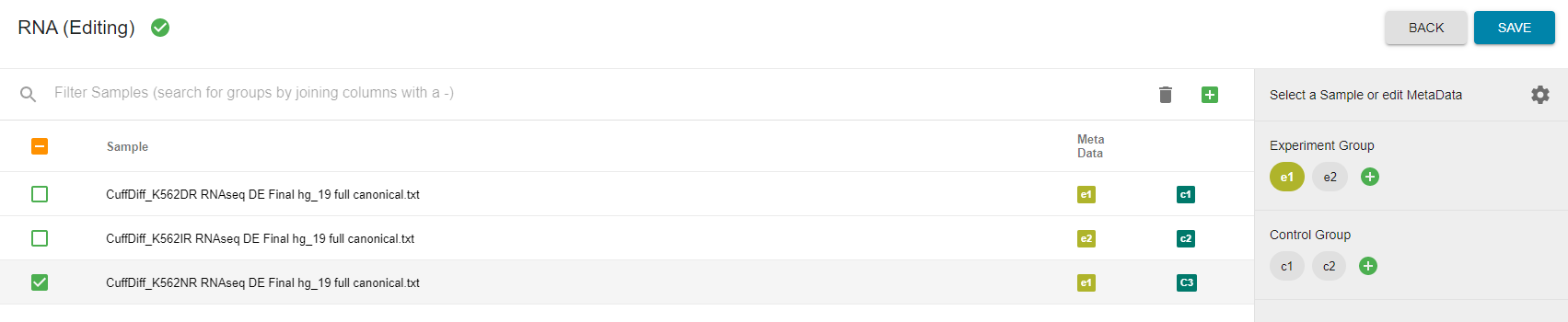
Now your transcriptomics data can be used in BioReviews and compared with your quantitative proteomics and metabolomics data.
RUO-MKT-18-12201-A



 Contact Support
Contact Support
0 Comments