The “Intensity (Peptide)” values come from LCMSReconstruct, in ProteinPilot software 5.0. It maps the RT, m/z, intensity MS1 surface to find the peak information for the peptide. The Intensity (Peptide) is a weighted sum of the heights of the isotope series at the...
Tags
Pharma perspectives: The influence of LC-MS innovation on drug development outsourcing
It is no secret that (bio)pharmaceutical research and development is complex, both scientific and regulatory processes. Working for a contract research organization and more recently for SCIEX has provided an interesting perspective on trends the market experiences that affect many of us.

Nitrosamines: Where are we now?
Nitrosamines are a large group of N-nitroso compounds that share a common functional N-N=O group. They are produced by a chemical reaction between a nitrosating agent and a secondary or tertiary amine. Back in 2018, nitrosamines suddenly found themselves in the spotlight when they were unexpectedly detected in medications for high blood pressure. Since then, they have been found in several other prescription medications, including those for heartburn, acid reflux and diabetes, resulting in manufacturers recalling some common medications.
High mass tuning calibration for ZenoTOF 7600
Hi,
Automation integration for the Echo® MS system
The Echo® MS system is specifically designed to be compatible with a variety of automation options to allow labs the flexibility to personalize their setup to meet their specific needs. To help you make the best decisions for your own lab, here are the answers to some...
Tips to maximize electrode lifetime for Echo MS system
While it’s easy to think of the Echo® MS system as an ultrafast LC system in front of the SCIEX Triple Quad 6500+ mass spectrometer, the system operates on fundamentally different principles. For this reason, it requires different routine maintenance to keep it...
Clustering algorithms in MarkerView App
Finding interesting and significant changes across a biological dataset can be challenging, so having good algorithms that can mine large datasets for differences, and find the statistically significant changes is really important. Within the MarkerView App we have...
How do I define the experimental design (the metadata) for my SWATH acquisition study within the OneOmics suite? What are the requirement for replicates?
In quantitative Omics research, the goal is to understand which analytes (protein or metabolite) are perturbed between experimental conditions; therefore we carefully design our studies to explore these questions. The algorithms used within the Assembler application...
What is the Most likely ratio normalization strategy?
When performing LC-MS quantitation, there are numerous sources of experimental variance that can confound the quality of your results (variation in the starting amount of sample, variation in the LC-MS measurements, etc.). Having a robust normalization strategy that...
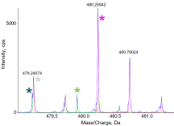
Understanding MS1 Peak Intensity in ProteinPilot software
The “Intensity (Peptide)” values come from LCMSReconstruct, in ProteinPilot software 5.0. It maps the RT, m/z, intensity MS1 surface to find the peak information for the peptide. The Intensity (Peptide) is a weighted sum of the heights of the isotope series at the...
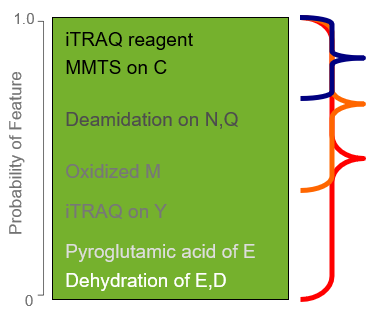
What is the difference between a rapid and a thorough search in ProteinPilot software?
When setting up your search in ProteinPilot software, you select either a Rapid Search or a Thorough Search in the Search Effort section. This setting determines which parts of the algorithm will be invoked and effectively how deep into your sample you will search to...
No Results Found
The page you requested could not be found. Try refining your search, or use the navigation above to locate the post.
No Results Found
The page you requested could not be found. Try refining your search, or use the navigation above to locate the post.