Hi,
Tags
Excel macro for calculating averages or percent CVs from replicates in a 384 -well plate
This excel workbook is designed to be used with data copied from a SCIEX OS software Analytics module results file

Back to the new basics: Part 1 | Making the leap from GC-MS to LC-MS
Producing accurate results quickly in a demanding environment is no easy feat for analytical scientists. What’s more, many of us are constantly questioning ourselves—I certainly am—about whether we are employing the best technique for the analysis at hand.
It’s an overwhelming thought, considering the wide range of tools that are available to choose from, each of which offers varying levels of capacity, sensitivity, selectivity, specificity and cost. How do you meet the unique needs of your organization without breaking the bank? I get it, and I’m not here to convince you it’s easy. My aim is to guide you through the process to help you make the right decision for you.
How to optimize sample plating to run Echo MS system in ‘Fast acquisition mode’
The Echo MS system can acquire data extremely rapidly, at 1 second per sample. To achieve this speed, it should be ensured that there is adequate time between ejections for the analyte signal to return to baseline between ejections. Plating your samples such that you...
How do I check the quality of the Auto Retention Time Calibration used in my Extractor processing?
When using the Auto-Calibration option in Extractor, a set of retention time calibration peptides will be determined automatically from the library and used for RT calibration. To determine how the fit looks for the calibration on each datafile, follow these steps....
sMRM Pro Builder template tutorial
The sMRM Pro Builder template is an Excel-based tool that can help you implement large panels of analytes in your lab. The Excel sheet will take your preliminary experimental results and compute retention times, retention time window widths and dwell time weighting to optimize your targeted assay.
MRM method transfer from a SCIEX Triple Quad or QTRAP 6500+ system to the SCIEX 7500 system
General recommendations when beginning method development Objective: The purpose of this document is to provide a quick reference for transferring MRM-based quantification methods from a SCIEX Triple Quad or QTRAP 6500+ system to a SCIEX 7500 system. While the best...
Short-chain PFAS compounds are on the rise- Craig’s PFAS Vodcast Cora Young
Read time: 2 minutes Short-chain per- and polyfluoroalkyl substances (PFAS) are increasing in the Canadian Arctic environment, with the most rapid increases occurring post-2000, according to a recent study in Geophysical Research Letters (April 2020). For example,...

Identifying the unknown PFAS profile in firefighting foams/AFFF
According to a recent study from Harvard University, the US EPA, and NIEHS, traditional targeted analysis techniques poorly characterize the PFAS composition of contemporary PFAS-based firefighting foams, know as aqueous film-forming foams (AFFF). Using the EPA 533 PFAS drinking water method for the analyte list, the researchers found that targeted mass spectrometry methods accounted for <1% of organic fluorine content. This is important because it demonstrates that targeted analysis methods miss nearly all the PFAS compounds in modern AFFF mixtures, thus underestimating the risk to human health and the environment.
sMRM Concurrency Calculator
This excel sheet allows you to quickly estimate the MRM concurrency and approximate dwell time for your Scheduled MRM Algorithm acquisition method. Paste your MRM method with retention times into the input tab then calculate the Excel workbook. Plots will update to...
Stay informed on the latest novel psychoactive substances (NPS) flooding the US drug market
Each year, the Drug Enforcement Administration (DEA) Special Testing and Research Laboratory publishes an Emerging Threat Report. The data contained in the report is updated quarterly and represents a snapshot of the drug evidence seized and analyzed by the DEA in the...
How do I define the experimental design (the metadata) for my SWATH acquisition study within the OneOmics suite? What are the requirement for replicates?
In quantitative Omics research, the goal is to understand which analytes (protein or metabolite) are perturbed between experimental conditions; therefore we carefully design our studies to explore these questions. The algorithms used within the Assembler application...
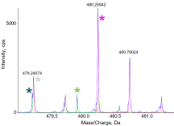
Understanding MS1 Peak Intensity in ProteinPilot software
The “Intensity (Peptide)” values come from LCMSReconstruct, in ProteinPilot software 5.0. It maps the RT, m/z, intensity MS1 surface to find the peak information for the peptide. The Intensity (Peptide) is a weighted sum of the heights of the isotope series at the...
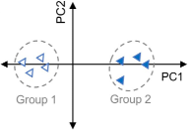
What are my normalization options in MarkerView software and when should I use them?
In an LC-MS experiment there are multiple sources of variance that can confound the quality of your results. This variation can be biological e.g. differences between treated and control groups, but can also be non-biological, usually from small variations in...
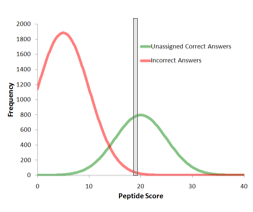
Computing protein confidence with improved accuracy by reassessing peptide confidence during protein grouping
ProteinPilot software 4.0 and higher releases include a new method for calculating protein confidences that improves reliability at the end of protein lists. Figure 1 shows a simulated example that demonstrates how more accurate protein confidence is computed. Figure...
Intabio acquisition expands SCIEX portfolio of impactful solutions to accelerate biotherapeutic development
Due to the nature of their production, biotherapeutics are difficult to manufacture. Growth conditions, purification protocols and formulation requirements can introduce unintended modifications into the protein structure that may affect its efficacy and safety.

Breaking down the SCIEX Triple Quad™ 7500 LC-MS/MS System – QTRAP® Ready
Sensitivity and robustness carry different meanings in the world of mass spectrometry. Generally, sensitivity refers to an instrument’s ability to achieve lower limits of detection (LOD). Robustness, on the other hand, refers to an instrument’s ability to consistently...
The honey sting
As a consumer it’s hard for me not to feel inundated with claims that our food is “all-natural” or “chemical-free” or that we should buy certain “superfoods” for their health benefits. We read labels and trust that the product we are buying is what we are truly...

Innovation that’s blasting through limitations in explosive detection
Mass spectrometry’s important role in identifying explosives The need for rapid explosive detection is now an unfortunate reality. The remit is multifaceted. The first is for preventative purposes, to protect us from any threat to life. The second is in the...
A new generation of therapeutic modalities
There are over 7,000 genetic diseases that could potentially be cured using gene therapy. Rare metabolic diseases, autoimmune disorders, cardiovascular disease and cancers are some of the top disease classes that can be addressed with gene therapies. With over 1,000...
Enhancing Biologics with CESI-MS Characterization
Comprehensive characterization of a biologic requires analysis at both the intact and digest levels, but these analyses can be complex and cumbersome. For example, with conventional liquid chromatography separations, researchers are often left with limited information...
Full, partial and empty capsid ratios for AAV analysis: What’s the big deal?
For many of you working to develop gene therapy drugs, you know that the time to market the drug is critical. Because gene therapeutics cure diseases by targeting specific genes, it is a constant race to see who develops the drug first. Unlike other classes of drugs where multiple medications can be used to treat a disease, whoever is first to develop a gene therapy drug wins.
Accurate mass LC-MS/MS for PFAS analysis without needing a blockbuster budget
If you’ve been following our recent blogs, you’ve probably seen quite a bit on how per- and polyfluoroalkyl substances (PFAS) are shaking up both the food and environmental industry. Even if you’ve not been following our blogs, you’ve probably seen a lot of media...

A rising star in food allergen research: proteomics of shellfish allergen
It’s important to know what you’re eating, especially if you suffer from a food allergy.
About 220 million people worldwide live with a food allergy.1 These numbers, along with the complexity and severity of conditions, continue to rise. In America, there are about 32 million food allergy sufferers—5.6 million of those are children under the age of 18.2.2 That’s 1 out of every 13 children, or about 2 in every classroom. From a financial perspective, the cost of food allergy childcare for US families is up to $25 billion

Routine cannabis screening is here. Will your lab reap the benefits?
Fast, accurate, and robust solution for routine commercial cannabis testing
As the world debates cannabis legalization for therapeutic applications and recreational use, the trends are shifting. Medicinal use of cannabis is legal in an increasing number of countries worldwide, including 33 states and the District of Columbia in the United States. Uruguay was the first country in the world to legalize the sale, cultivation, and distribution of cannabis in 2013. In the United States, Washington and Colorado were the first to fully legalize cannabis in 2012. By the end of 2019, 10 states have legalized recreational use for adults over the age of 21, with 64% of Americans favoring the move
Keep up with fentanyl and its analogs: Failure to do so is not an option.
Fentanyl is one of the most well-known synthetic opioids that has been in the spotlight lately—and not for good reasons. The World Drug Report 2019 reveals that there have been more than 47,000 opioid overdose deaths in the United States in 2017. This is an increase...

Hemp, marijuana, CBD and THC: what’s the difference?
Cannabis refers to a genus of flowering plants originating from Asia.1 It's also an umbrella term that includes both marijuana plants and hemp plants, among others. With the flurry of legalization across the United States, CBD and THC derived products have been thrown...

Mysterious Vaping Detectives Part 2
In our last blog, I gave you some current highlights about the mysterious vaping illness that was making headlines (and still is). The condition now known as, e-cigarette or vaping, product use-associated lung injury (EVALI) has sickened thousands and killed 57...
Trends shaping global environmental analytical testing
The only certainty in a changing environmental landscape Despite a 38-fold increase in environmental laws put in place around the world since 1972, the future of our planet is under the spotlight like never before. Mitigating climate change has arrived as our world's...

Mass spectrometry as a forensic science tool in 2020
There’s no doubt about it: forensics is at the front line of the criminal justice system. It’s where analytical chemistry has the power to fight crimes. It’s fascinating! But what excites me is how quickly things change and the speed of innovation. Just think about...
Microflow for metabolite ID: a win all around
Over the last several years there has been a slow and steady progression within the LC-MS community to move traditional high-flow applications to lower flow rates. In particular, moving into the microflow regime has proven to be a simple adjustment in methodology that...



 Contact Support
Contact Support